
The generation of distinctive cell types that form different tissues and organs requires precise, temporal and spatial control of gene expression. This depends on specific cis-regulatory elements distributed in the non-coding DNA surrounding their target genes. Studies performed on mammalian embryonic stem cells and Drosophila embryos suggest that active enhancers form part of a defined chromatin landscape marked by histone H3 lysine 4 mono-methylation (H3K4me1) and histone H3 lysine 27 acetylation (H3K27ac). Nevertheless, little is known about the dynamics and the potential roles of these marks during vertebrate embryogenesis. We have provided genomic maps of H3K4me1/me3 and H3K27ac at four developmental time-points of zebrafish embryogenesis and analyze embryonic enhancer activity.
We have found that:
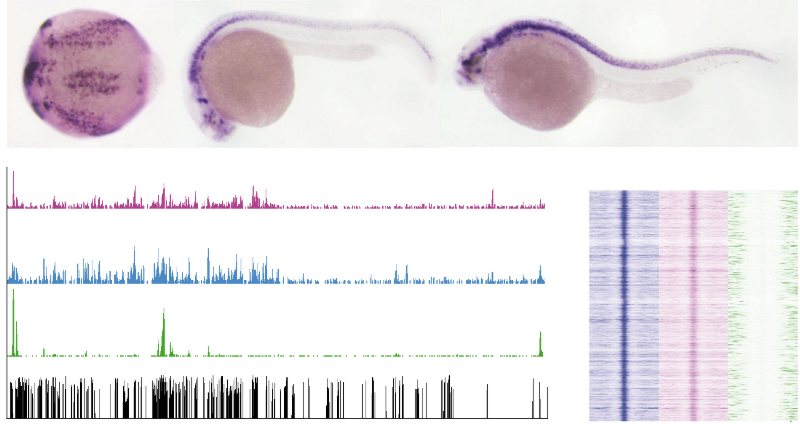
Our published data, together with public available RNA-seq, can be visualized at ZV9 public.
Our published data can be downloaded from:
GEO H3K4me1 , H3K4me3 , H3K27ac ,
GEO H3K27me3.