
Using Next Generation Sequencing (NGS), we are analyzing how different epigenomic signatures change as vertebrate embryogenesis progresses. By genome-wide interrogation of histone marks associated with enhancers and promoters in zebrafish, we determined, for the first time in a vertebrate embryo, the dynamics of enhancer activity during early embryogenesis. We are extending these studies to other histone and DNA modifications associated with repressive states. We aim to integrate all these data to better understand the chromatin modifications associated to differentiation processes that shape the early embryo. The published data can be visualized at Epigenomics page.
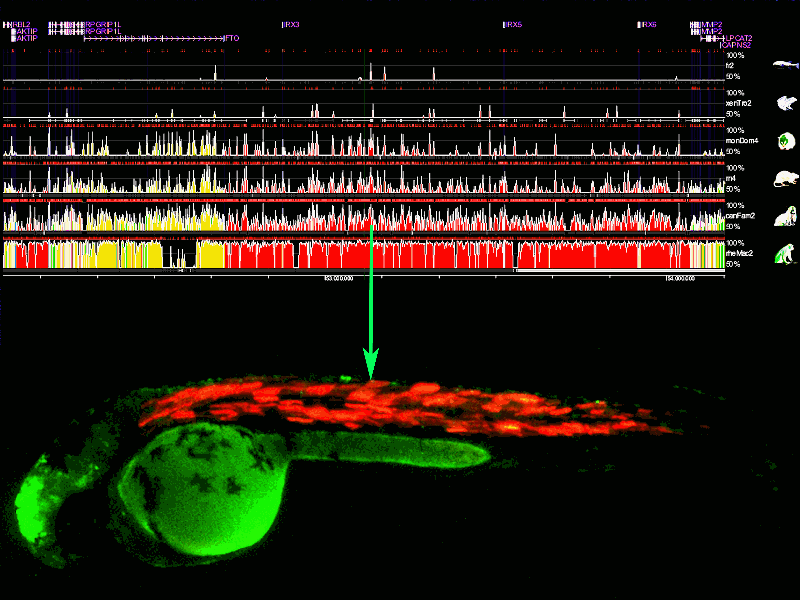
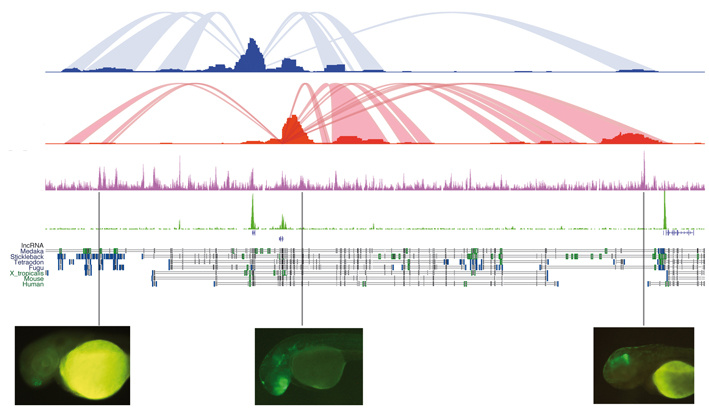
Epigenomic information has facilitated the identification, in a genome-wide manner, of potential cis-regulatory elements (CREs) active in a particular cell type, tissue or embryonic stage. The next required step is to identify the target genes of all these immense collections of CREs. The DNA containing all CREs acting on a particular gene has been denominated the regulatory landscape of the gene in question. These landscapes can be very large, especially for developmental genes that are controlled by multiple regulatory regions that can be localized in genomic intervals of megabase sizes. A recently developed technique (4C-seq), based on NGS, allows for effective identification of the regulatory landscapes of genes. We are applying this technique to systematically determine the gene regulatory landscapes of key developmental genes in embryos of different species.
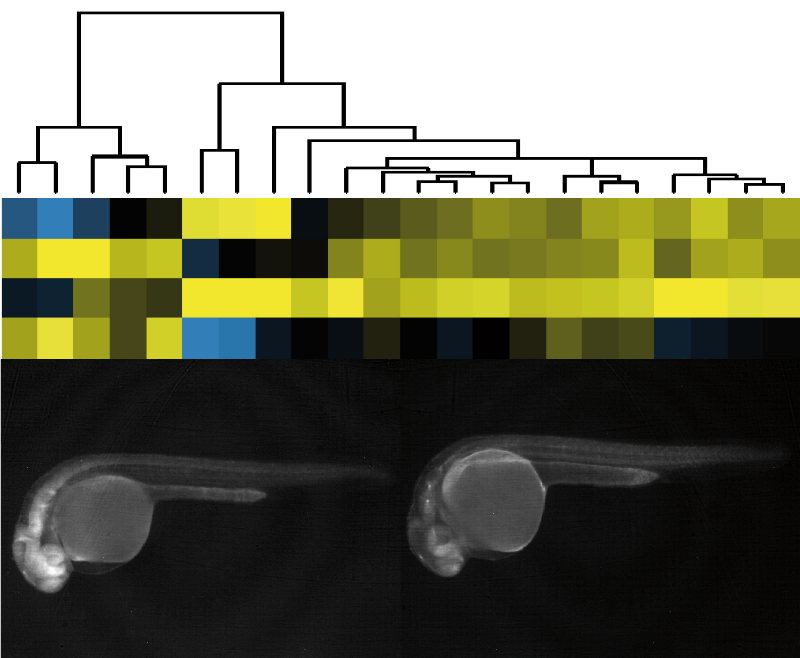
We have recently designed the Expression Disruption (ED) vector. ED is an engineered Tol2 transposon that includes a strong insulator flanked by two fluorescent reporter genes under the control of minimal promoters that function as independent enhancer traps. The strong insulator in the ED transposon is expected to disrupt the regulatory landscapes in which it becomes integrated. If this disruption is effective, the regulatory information upstream or downstream of the integration site should differentially activate Green Fluorescent Protein (GFP) or Red Fluorescent Protein (RFP) reporters, positioned on either side of the insulator. We have generated hundreds of zebrafish ED lines which can be visualized at EDscreen's page. A detailed evaluation of a fraction of these lines shows that at least half of them disrupt cis-regulatory landscapes causing alterations on gene expression and function. We are currently generating new tissue-specific lines.
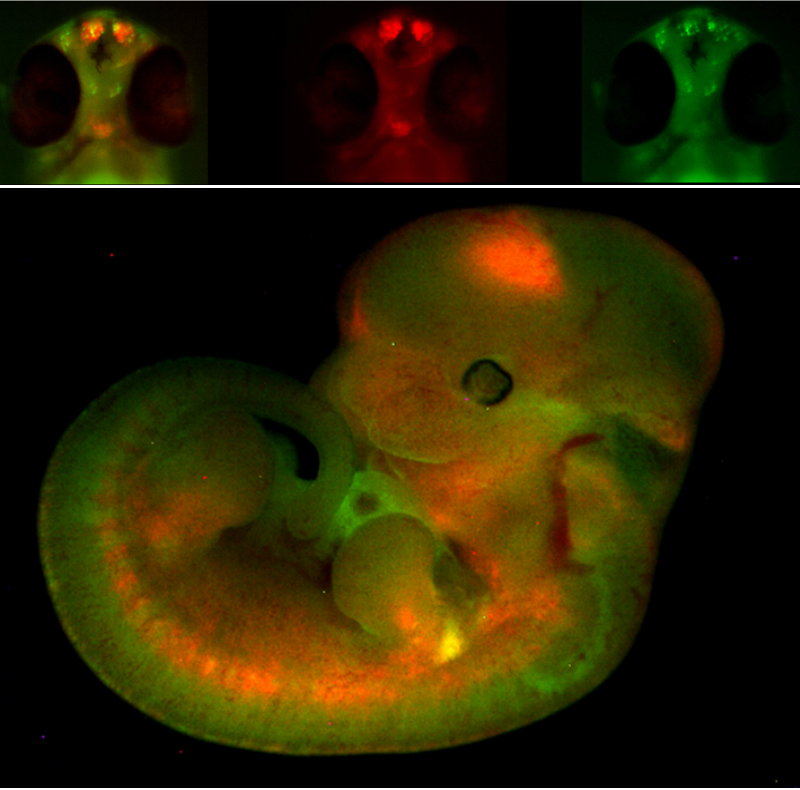
We are interested in cis-regulatory elements that have important function in evolution. In the recent years, we have functionally tested, through stable transgenic assays in zebrafish, enhancer activity of many Highly Conserved Non-coding Regions. Pictures of these lines can be found at Enhancers page. Currently, we are combining ChIP-seq, 4C-seq and enhancer transgenic assays to identify CREs with key function in evolution.
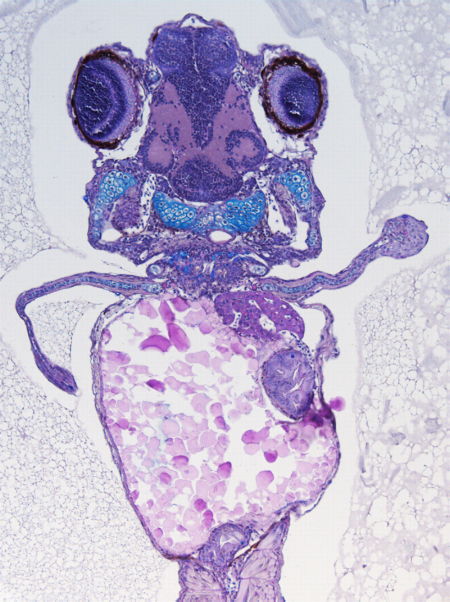
Through multiple collaborations with different human genetics groups, we have and we are evaluating the functional impact of human mutation in the activity of different type of cis-regulatory elements.